xarray¶
multi-dimensional data analysis in Python¶
ACINN workshop, Tue 07.02.2017
Fabien Maussion

Slides: http://fabienmaussion.info/acinn_xarray_workshop
Notebook: On GitHub
xarray¶

Documentation: http://xarray.pydata.org
Repository: https://github.com/pydata/xarray
Initial release: 03.05.2014
Latest release: v0.9.1 (20.01.2017)
53 contributors (latest release: 24)
Umbrellas: Python for data & NumFOCUS (but no funding...)


numpy.array¶
import numpy as np
a = np.array([[1, 3, 9], [2, 8, 4]])
a
array([[1, 3, 9],
[2, 8, 4]])
a[1, 2]
4
a.mean(axis=0)
array([ 1.5, 5.5, 6.5])
xarray.DataArray¶
import xarray as xr
da = xr.DataArray(a, dims=['lat', 'lon'],
coords={'lon':[11, 12, 13], 'lat':[1, 2]})
da
<xarray.DataArray (lat: 2, lon: 3)>
array([[1, 3, 9],
[2, 8, 4]])
Coordinates:
* lon (lon) int64 11 12 13
* lat (lat) int64 1 2
da.sel(lon=13, lat=2).values
array(4)
da.mean(dim='lat')
<xarray.DataArray (lon: 3)> array([ 1.5, 5.5, 6.5]) Coordinates: * lon (lon) int64 11 12 13
xarray.Dataset¶
f = 'ERA-Int-MonthlyAvg-4D-TUVWZ.nc'
ds = xr.open_dataset(f)
ds
<xarray.Dataset>
Dimensions: (latitude: 241, level: 15, longitude: 480, month: 12)
Coordinates:
* latitude (latitude) float32 90.0 89.25 88.5 87.75 87.0 ...
* level (level) int32 50 100 150 200 300 400 500 600 ...
* longitude (longitude) float32 -180.0 -179.25 -178.5 ...
* month (month) int64 1 2 3 4 5 6 7 8 9 10 11 12
Data variables:
u (month, level, latitude, longitude) float64 10.38 ...
v (month, level, latitude, longitude) float64 5.594 ...
w (month, level, latitude, longitude) float64 -0.0003052 ...
z (month, level, latitude, longitude) float64 1.888e+05 ...
t (month, level, latitude, longitude) float64 201.1 ...
Attributes:
Conventions: CF-1.0
Info: Monthly ERA-Interim data. Downloaded and edited by fabien.maussion@uibk.ac.at
Selection¶
By value¶
ds.t.sel(month=8, level=850)
<xarray.DataArray 't' (latitude: 241, longitude: 480)>
[115680 values with dtype=float64]
Coordinates:
* latitude (latitude) float32 90.0 89.25 88.5 87.75 87.0 ...
level int32 850
* longitude (longitude) float32 -180.0 -179.25 -178.5 ...
month int64 8
Attributes:
units: K
long_name: Temperature
standard_name: air_temperature
By index¶
ds.t.isel(month=7, level=11)
<xarray.DataArray 't' (latitude: 241, longitude: 480)>
[115680 values with dtype=float64]
Coordinates:
* latitude (latitude) float32 90.0 89.25 88.5 87.75 87.0 ...
level int32 850
* longitude (longitude) float32 -180.0 -179.25 -178.5 ...
month int64 8
Attributes:
units: K
long_name: Temperature
standard_name: air_temperature
By "wait... where is Innsbruck again?"¶
ds.t.sel(level=1001, latitude=47.26, longitude=11.38, method='nearest')
<xarray.DataArray 't' (month: 12)>
array([ 278.921875, 279.09375 , 282.140625, 285.359375,
290.132812, 293.390625, 295.84375 , 295.867188,
292.21875 , 288.484375, 283.117188, 280.054688])
Coordinates:
latitude float32 47.25
level int32 1000
longitude float32 11.25
* month (month) int64 1 2 3 4 5 6 7 8 9 10 11 12
Attributes:
units: K
long_name: Temperature
standard_name: air_temperature
The "old way"¶
ds.t[7, 11, :, :]
<xarray.DataArray 't' (latitude: 241, longitude: 480)>
[115680 values with dtype=float64]
Coordinates:
* latitude (latitude) float32 90.0 89.25 88.5 87.75 87.0 ...
level int32 850
* longitude (longitude) float32 -180.0 -179.25 -178.5 ...
month int64 8
Attributes:
units: K
long_name: Temperature
standard_name: air_temperature
Operations¶
Aggregation¶
ds.u.mean(dim=['month', 'longitude']).plot.contourf(levels=13)
plt.ylim([1000, 100]);
And other kind of things¶
u_avg = ds.u.mean(dim=['month', 'longitude'])
u_avg_masked = u_avg.where(u_avg > 12)
u_avg_masked.plot.contourf(levels=13)
plt.ylim([1000, 100]);
Arithmetic¶
Broadcasting¶
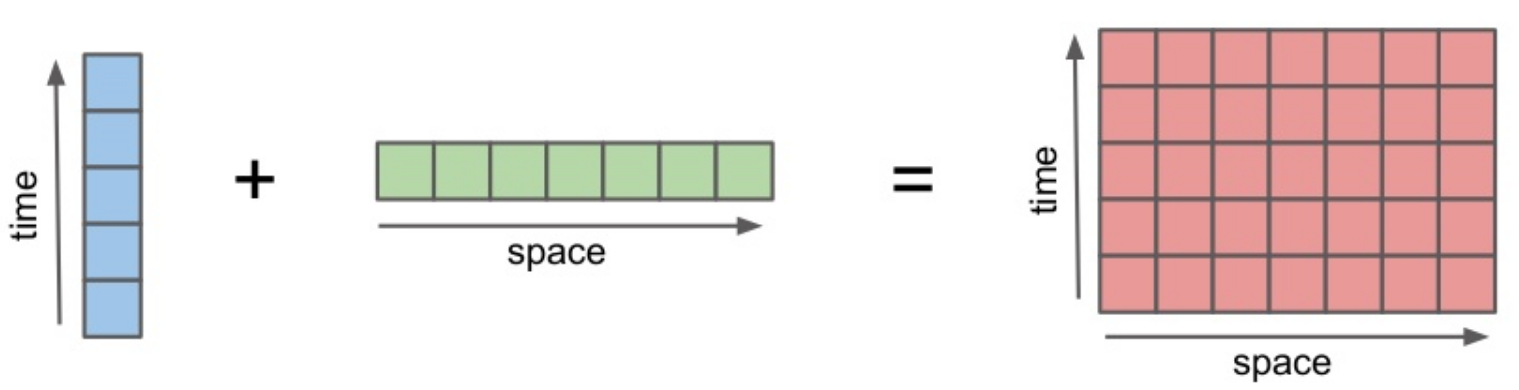
a = xr.DataArray(np.arange(3), dims='time',
coords={'time':np.arange(3)})
b = xr.DataArray(np.arange(4), dims='space',
coords={'space':np.arange(4)})
a + b
<xarray.DataArray (time: 3, space: 4)>
array([[0, 1, 2, 3],
[1, 2, 3, 4],
[2, 3, 4, 5]])
Coordinates:
* time (time) int64 0 1 2
* space (space) int64 0 1 2 3
Alignment¶
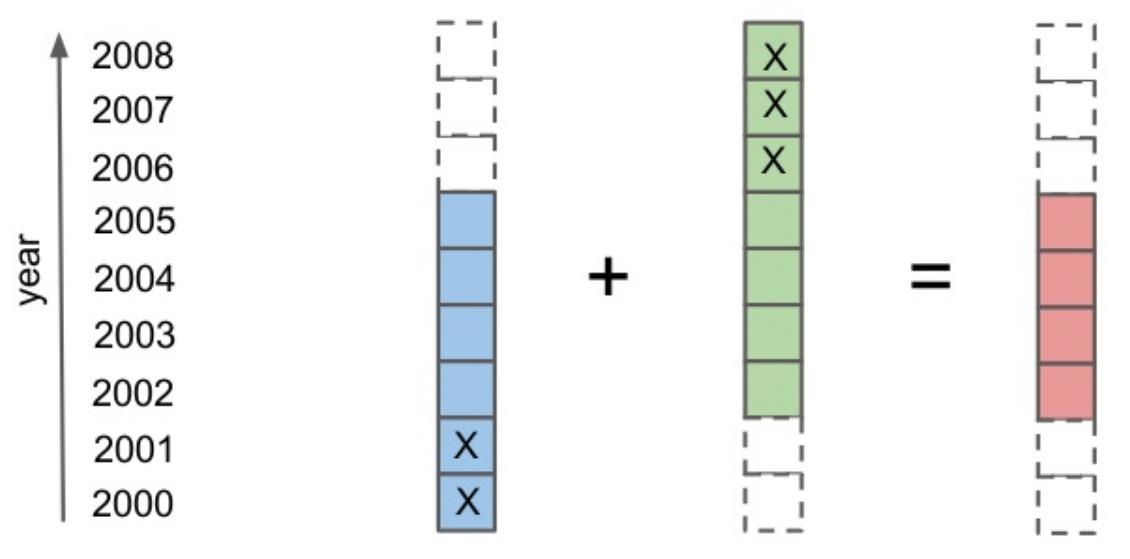
a = xr.DataArray(np.arange(3), dims='time',
coords={'time':np.arange(3)})
b = xr.DataArray(np.arange(5), dims='time',
coords={'time':np.arange(5)+1})
a + b
<xarray.DataArray (time: 2)> array([1, 3]) Coordinates: * time (time) int64 1 2
Plotting¶
1-d¶
ts = ds.t.sel(level=1001, latitude=47.26, longitude=11.38, method='nearest')
ts.plot();
On maps¶
import cartopy.crs as ccrs
ax = plt.axes(projection=ccrs.Robinson())
ds.z.sel(level=1000, month=8).plot(ax=ax, transform=ccrs.PlateCarree());
ax.coastlines();
(Big) data: multiple files¶
Opening all files in a directory...
mfs = '/home/mowglie/disk/Data/Gridded/GPM/3BDAY_sorted/*.nc'
dsmf = xr.open_mfdataset(mfs)
... results in a consolidated dataset ...
dsmf
<xarray.Dataset>
Dimensions: (lat: 732, lon: 620, time: 672)
Coordinates:
* lat (lat) float32 -56.95 -56.85 -56.75 ...
* lon (lon) float32 -93.55 -93.45 -93.35 ...
* time (time) datetime64[ns] 2014-03-31 ...
Data variables:
precipitationCal (time, lat, lon) float64 0.0 0.0 0.3 ...
precipitationHQ (time, lat, lon) float64 0.0 0.0 0.325 ...
... on which all usual operations can be applied:
dsmf = dsmf.sel(time='2015')
dsmf
<xarray.Dataset>
Dimensions: (lat: 732, lon: 620, time: 365)
Coordinates:
* lat (lat) float32 -56.95 -56.85 -56.75 ...
* lon (lon) float32 -93.55 -93.45 -93.35 ...
* time (time) datetime64[ns] 2015-01-01 ...
Data variables:
precipitationCal (time, lat, lon) float64 0.0 0.0 0.0 ...
precipitationHQ (time, lat, lon) float64 0.0 0.0 0.0 ...
Yes, even computations!
ts = dsmf.precipitationCal.mean(dim=['lon', 'lat'])
ts
<xarray.DataArray 'precipitationCal' (time: 365)> dask.array<mean_ag..., shape=(365,), dtype=float64, chunksize=(1,)> Coordinates: * time (time) datetime64[ns] 2015-01-01 2015-01-02 ...
Computations are done "lazily"
No actual computation has happened yet:
ts.data
dask.array<mean_ag..., shape=(365,), dtype=float64, chunksize=(1,)>
But they can be triggered:
ts = ts.load()
ts
<xarray.DataArray 'precipitationCal' (time: 365)>
array([ 2.297214, 3.00098 , 2.532836, ..., 2.516468,
2.334409, 3.469001])
Coordinates:
* time (time) datetime64[ns] 2015-01-01 2015-01-02 ...
For more information: http://xarray.pydata.org/en/stable/dask.html
ts.plot();
ts.rolling(time=31, center=True).mean().plot();
Extensions¶
Example: EOFS¶
Taken from: http://ajdawson.github.io/eofs/examples/nao_xarray.html
from eofs.xarray import Eof
from eofs.examples import example_data_path
# Read geopotential height data using the xarray module
filename = example_data_path('hgt_djf.nc')
z_djf = xr.open_dataset(filename)['z']
# Compute anomalies by removing the time-mean.
z_djf = z_djf - z_djf.mean(dim='time')
# Create an EOF solver to do the EOF analysis.
coslat = np.cos(np.deg2rad(z_djf.coords['latitude'].values)).clip(0., 1.)
solver = Eof(z_djf, weights=np.sqrt(coslat)[..., np.newaxis])
# Get the leading EOF
eof1 = solver.eofsAsCovariance(neofs=1)
# Leading EOF expressed as covariance in the European/Atlantic domain
ax = plt.axes(projection=ccrs.Orthographic(central_longitude=-20, central_latitude=60))
ax.coastlines() ; ax.set_global()
eof1[0, 0].plot.contourf(ax=ax, levels=np.linspace(-75, 75, 11),
cmap=plt.cm.RdBu_r, add_colorbar=False,
transform=ccrs.PlateCarree())
ax.set_title('EOF1 expressed as covariance', fontsize=16);
Salem¶
- Adds geolocalized operations to xarray
- Adds projection transformations
- Adds WRF support
http://salem.readthedocs.io/en/latest/
Try it out:
pip install salemPlotting¶
# importing salem adds a new "toolbox" to xarray objects
import salem
pday = dsmf.precipitationCal.sel(time='2015-02-01')
cm = pday.salem.quick_map(cmap='Blues', vmax=100);
Subsetting¶
shdf = salem.read_shapefile(salem.get_demo_file('world_borders.shp'))
shdf = shdf.loc[shdf['CNTRY_NAME'].isin(['Peru'])]
dsmfperu = dsmf.salem.subset(shape=shdf, margin=10)
pday = dsmfperu.precipitationCal.sel(time='2015-02-01')
cm = pday.salem.quick_map(cmap='Blues', vmax=100);
Regions of interest¶
dsmfperu = dsmfperu.salem.roi(shape=shdf)
pday = dsmfperu.precipitationCal.sel(time='2015-02-01')
cm = pday.salem.quick_map(cmap='Blues', vmax=100);
... all xarray operations continue to apply¶
prpc_a = dsmfperu.precipitationCal.sum(dim=['time']).load()
prpc_a.salem.quick_map(cmap='Blues', vmax=5000);
WRF output files¶
Problems:
- not CF compliant (e.g. timestamp)
- staggered grids
- not all variables available (e.g. moisture transport)
- large
Example file¶
f = 'wrfpost_d01_2005-09-21_00-00-00_25h.nc'
ds = xr.open_dataset(f)
ds
<xarray.Dataset>
Dimensions: (Time: 9, bottom_top: 27, bottom_top_stag: 28, soil_layers_stag: 4, south_north: 200, south_north_stag: 201, west_east: 200, west_east_stag: 201)
Coordinates:
XLAT (south_north, west_east) float32 1.62003 ...
XLAT_U (south_north, west_east_stag) float32 1.59409 ...
XLAT_V (south_north_stag, west_east) float32 1.50526 ...
XLONG (south_north, west_east) float32 63.3768 ...
XLONG_U (south_north, west_east_stag) float32 63.262 ...
XLONG_V (south_north_stag, west_east) float32 63.4026 ...
Dimensions without coordinates: Time, bottom_top, bottom_top_stag, soil_layers_stag, south_north, south_north_stag, west_east, west_east_stag
Data variables:
Times (Time) |S19 b'2005-09-21_00:00:00' ...
ACGRDFLX (Time, south_north, west_east) float32 0.0 ...
ACHFX (Time, south_north, west_east) float32 173706.0 ...
ACLHF (Time, south_north, west_east) float32 5.62606e+06 ...
ALBBCK (Time, south_north, west_east) float32 0.08 ...
ALBEDO (Time, south_north, west_east) float32 0.08 ...
AVG_FUEL_FRAC (Time, south_north, west_east) float32 0.0 ...
CANWAT (Time, south_north, west_east) float32 0.0 ...
CF1 (Time) float32 1.76177 1.76177 ...
CF2 (Time) float32 -1.02047 -1.02047 ...
CF3 (Time) float32 0.258698 0.258698 ...
CFN (Time) float32 1.46268 1.46268 ...
CFN1 (Time) float32 -0.462685 -0.462685 ...
CLDFRA (Time, bottom_top, south_north, west_east) float32 0.0 ...
COSALPHA (south_north, west_east) float32 0.975546 ...
DN (bottom_top) float32 0.0 ...
DNW (bottom_top) float32 -0.00700003 ...
DZS (soil_layers_stag) float32 0.1 0.3 ...
E (south_north, west_east) float32 0.000145782 ...
EL_PBL (Time, bottom_top, south_north, west_east) float32 0.0 ...
EMISS (Time, south_north, west_east) float32 0.98 ...
F (south_north, west_east) float32 4.12304e-06 ...
FNM (bottom_top) float32 0.0 0.411766 ...
FNP (bottom_top) float32 0.0 0.588234 ...
GLW (Time, south_north, west_east) float32 404.028 ...
GRAUPELNC (Time, south_north, west_east) float32 0.0 ...
GRDFLX (Time, south_north, west_east) float32 0.0 ...
HAILNC (Time, south_north, west_east) float32 0.0 ...
HFX (Time, south_north, west_east) float32 4.29665 ...
HFX_FORCE (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
HFX_FORCE_TEND (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
HGT (south_north, west_east) float32 0.0 ...
H_DIABATIC (Time, bottom_top, south_north, west_east) float32 0.0 ...
ISLTYP (south_north, west_east) int32 14 ...
ITIMESTEP (Time) int32 288 360 432 504 576 ...
IVGTYP (south_north, west_east) int32 16 ...
K22_SHALLOW (Time, south_north, west_east) int32 0 ...
KBCON_SHALLOW (Time, south_north, west_east) int32 0 ...
KTOP_SHALLOW (Time, south_north, west_east) int32 0 ...
LAI (south_north, west_east) float32 0.0 ...
LANDMASK (south_north, west_east) float32 0.0 ...
LH (Time, south_north, west_east) float32 140.399 ...
LH_FORCE (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
LH_FORCE_TEND (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
LU_INDEX (south_north, west_east) float32 16.0 ...
MAPFAC_M (south_north, west_east) float32 1.14636 ...
MAPFAC_MX (south_north, west_east) float32 1.14636 ...
MAPFAC_MY (south_north, west_east) float32 1.14636 ...
MAPFAC_U (south_north, west_east_stag) float32 1.14663 ...
MAPFAC_UX (south_north, west_east_stag) float32 1.14663 ...
MAPFAC_UY (south_north, west_east_stag) float32 1.14663 ...
MAPFAC_V (south_north_stag, west_east) float32 1.14754 ...
MAPFAC_VX (south_north_stag, west_east) float32 1.14754 ...
MAPFAC_VY (south_north_stag, west_east) float32 1.14754 ...
MAX_MSTFX (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
MAX_MSTFY (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
MF_VX_INV (south_north_stag, west_east) float32 0.871432 ...
MU (Time, south_north, west_east) float32 540.09 ...
MUB (Time, south_north, west_east) float32 95000.0 ...
NEST_POS (south_north, west_east) float32 0.0 ...
NOAHRES (Time, south_north, west_east) float32 0.0 ...
OLR (Time, south_north, west_east) float32 285.326 ...
P (Time, bottom_top, south_north, west_east) float32 951.938 ...
P00 (Time) float32 100000.0 100000.0 ...
PB (Time, bottom_top, south_north, west_east) float32 99667.5 ...
PBLH (Time, south_north, west_east) float32 803.482 ...
PH (Time, bottom_top_stag, south_north, west_east) float32 0.0 ...
PHB (Time, bottom_top_stag, south_north, west_east) float32 0.0 ...
POTEVP (Time, south_north, west_east) float32 0.0 ...
PSFC (Time, south_north, west_east) float32 100959.0 ...
P_HYD (Time, bottom_top, south_north, west_east) float32 100618.0 ...
P_TOP (Time) float32 5000.0 5000.0 5000.0 ...
Q2 (Time, south_north, west_east) float32 0.0201797 ...
QCLOUD (Time, bottom_top, south_north, west_east) float32 0.0 ...
QFX (Time, south_north, west_east) float32 5.61596e-05 ...
QGRAUP (Time, bottom_top, south_north, west_east) float32 0.0 ...
QICE (Time, bottom_top, south_north, west_east) float32 0.0 ...
QNICE (Time, bottom_top, south_north, west_east) float32 0.0 ...
QNRAIN (Time, bottom_top, south_north, west_east) float32 0.0 ...
QRAIN (Time, bottom_top, south_north, west_east) float32 0.0 ...
QSNOW (Time, bottom_top, south_north, west_east) float32 0.0 ...
QVAPOR (Time, bottom_top, south_north, west_east) float32 0.0168769 ...
RAINC (Time, south_north, west_east) float32 0.0 ...
RAINNC (Time, south_north, west_east) float32 0.0 ...
RAINSH (Time, south_north, west_east) float32 0.0 ...
RDN (bottom_top) float32 0.0 -117.647 ...
RDNW (bottom_top) float32 -142.857 ...
RDX (Time) float32 3.33333e-05 ...
RDY (Time) float32 3.33333e-05 ...
RESM (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
SAVE_TOPO_FROM_REAL (Time) int32 0 0 0 0 0 0 0 0 0
SEAICE (Time, south_north, west_east) float32 0.0 ...
SEED1 (Time) int32 0 0 0 0 0 0 0 0 0
SEED2 (Time) int32 0 0 0 0 0 0 0 0 0
SFROFF (Time, south_north, west_east) float32 0.0 ...
SH2O (Time, soil_layers_stag, south_north, west_east) float32 1.0 ...
SINALPHA (south_north, west_east) float32 0.219797 ...
SMCREL (Time, soil_layers_stag, south_north, west_east) float32 1.0 ...
SMOIS (Time, soil_layers_stag, south_north, west_east) float32 1.0 ...
SNOPCX (Time, south_north, west_east) float32 0.0 ...
SNOW (Time, south_north, west_east) float32 0.0 ...
SNOWC (Time, south_north, west_east) float32 0.0 ...
SNOWH (Time, south_north, west_east) float32 0.0 ...
SNOWNC (Time, south_north, west_east) float32 0.0 ...
SOILTB (Time, south_north, west_east) float32 0.0 ...
SR (Time, south_north, west_east) float32 0.0 ...
SST (Time, south_north, west_east) float32 302.047 ...
SSTSK (Time, south_north, west_east) float32 0.0 ...
STEPAVE_COUNT (Time) int32 0 0 0 0 0 0 0 0 0
SWDOWN (Time, south_north, west_east) float32 0.0 ...
SWNORM (Time, south_north, west_east) float32 0.0 ...
T (Time, bottom_top, south_north, west_east) float32 0.658561 ...
T00 (Time) float32 290.0 290.0 290.0 ...
T2 (Time, south_north, west_east) float32 301.709 ...
TH2 (Time, south_north, west_east) float32 300.887 ...
TISO (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
TKE_PBL (Time, bottom_top, south_north, west_east) float32 0.105927 ...
TLP (Time) float32 50.0 50.0 50.0 50.0 ...
TMN (Time, south_north, west_east) float32 302.265 ...
TSK (Time, south_north, west_east) float32 302.047 ...
TSK_FORCE (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
TSK_FORCE_TEND (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
TSLB (Time, soil_layers_stag, south_north, west_east) float32 302.047 ...
U (Time, bottom_top, south_north, west_east_stag) float32 -2.01244 ...
U10 (Time, south_north, west_east) float32 -1.86241 ...
UAH (Time, south_north, west_east_stag) float32 0.0 ...
UDROFF (Time, south_north, west_east) float32 0.0 ...
UST (Time, south_north, west_east) float32 0.20173 ...
V (Time, bottom_top, south_north_stag, west_east) float32 6.57835 ...
V10 (Time, south_north, west_east) float32 6.27481 ...
VAH (Time, south_north_stag, west_east) float32 0.0 ...
VEGFRA (south_north, west_east) float32 0.0 ...
W (Time, bottom_top_stag, south_north, west_east) float32 0.0 ...
XICEM (Time, south_north, west_east) float32 0.0 ...
XLAND (Time, south_north, west_east) float32 2.0 ...
XMB_SHALLOW (Time, south_north, west_east) float32 0.0 ...
XTIME (Time) float32 720.0 900.0 1080.0 ...
ZETATOP (Time) float32 0.0 0.0 0.0 0.0 0.0 ...
ZNU (Time, bottom_top) float32 0.9965 ...
ZNW (Time, bottom_top_stag) float32 1.0 ...
ZS (soil_layers_stag) float32 0.05 ...
Attributes:
TITLE: OUTPUT FROM WRF V3.3.1 MODEL
START_DATE: 2005-09-21_00:00:00
SIMULATION_START_DATE: 2005-09-20_12:00:00
WEST-EAST_GRID_DIMENSION: 201
SOUTH-NORTH_GRID_DIMENSION: 201
BOTTOM-TOP_GRID_DIMENSION: 28
DX: 30000.0
DY: 30000.0
GRIDTYPE: C
DIFF_OPT: 1
KM_OPT: 4
DAMP_OPT: 3
DAMPCOEF: 0.2
KHDIF: 0.0
KVDIF: 0.0
MP_PHYSICS: 8
RA_LW_PHYSICS: 1
RA_SW_PHYSICS: 1
SF_SFCLAY_PHYSICS: 2
SF_SURFACE_PHYSICS: 2
BL_PBL_PHYSICS: 2
CU_PHYSICS: 5
SURFACE_INPUT_SOURCE: 1
SST_UPDATE: 1
GRID_FDDA: 0
GFDDA_INTERVAL_M: 0
GFDDA_END_H: 0
GRID_SFDDA: 0
SGFDDA_INTERVAL_M: 0
SGFDDA_END_H: 0
SF_URBAN_PHYSICS: 0
FEEDBACK: 1
SMOOTH_OPTION: 0
SWRAD_SCAT: 1.0
W_DAMPING: 0
MOIST_ADV_OPT: 1
SCALAR_ADV_OPT: 1
TKE_ADV_OPT: 1
DIFF_6TH_OPT: 2
DIFF_6TH_FACTOR: 0.12
OBS_NUDGE_OPT: 0
BUCKET_MM: -1.0
BUCKET_J: -1.0
PREC_ACC_DT: 0.0
OMLCALL: 0
ISFTCFLX: 0
ISHALLOW: 0
DFI_OPT: 0
SHCU_PHYSICS: 0
WEST-EAST_PATCH_START_UNSTAG: 1
WEST-EAST_PATCH_END_UNSTAG: 200
WEST-EAST_PATCH_START_STAG: 1
WEST-EAST_PATCH_END_STAG: 201
SOUTH-NORTH_PATCH_START_UNSTAG: 1
SOUTH-NORTH_PATCH_END_UNSTAG: 200
SOUTH-NORTH_PATCH_START_STAG: 1
SOUTH-NORTH_PATCH_END_STAG: 201
BOTTOM-TOP_PATCH_START_UNSTAG: 1
BOTTOM-TOP_PATCH_END_UNSTAG: 27
BOTTOM-TOP_PATCH_START_STAG: 1
BOTTOM-TOP_PATCH_END_STAG: 28
GRID_ID: 1
PARENT_ID: 1
I_PARENT_START: 1
J_PARENT_START: 1
PARENT_GRID_RATIO: 1
DT: 150.0
CEN_LAT: 30.0
CEN_LON: 87.0
TRUELAT1: 30.0
TRUELAT2: 35.0
MOAD_CEN_LAT: 30.0
STAND_LON: 87.0
POLE_LAT: 90.0
POLE_LON: 0.0
GMT: 12.0
JULYR: 2005
JULDAY: 263
MAP_PROJ: 1
MMINLU: USGS
NUM_LAND_CAT: 28
ISWATER: 16
ISLAKE: 28
ISICE: 24
ISURBAN: 1
ISOILWATER: 14
history: Thu Nov 10 22:29:11 CET 2011: runncl.sh /cfs/tool/jc_1.4test/utils//POST_wrf.ncl ./wrfout_d01_2005-09-21_00-00-00_24h.nc . 0 vars_stat.csv vars_del.csv
Thu Nov 10 22:29:08 2011: ncks -d Time,4,12 wrfout_d01_2005-09-20_12:00:00 wrfout_d01_2005-09-21_00-00-00_24h.nc
NCO: 20111110
CREATION_DATE: Thu Nov 10 22:29:11 CET 2011
SPINUP_INDEX: 0
END_INDEX: 8
Objectives¶
- "clean" the file to make it more appealing
- automatic projection parsing
- automatic unstaggering
- pressure-levels interpolation
- diagnostic variables
- ...
wrf = salem.open_wrf_dataset(f)
wrf
<xarray.Dataset>
Dimensions: (bottom_top: 27, soil_layers: 3, south_north: 200, time: 9, west_east: 200)
Coordinates:
lat (south_north, west_east) float32 1.62003 ...
lon (south_north, west_east) float32 63.3768 ...
* time (time) datetime64[ns] 2005-09-21 ...
* west_east (west_east) float64 -2.985e+06 ...
* south_north (south_north) float64 -2.985e+06 ...
Dimensions without coordinates: bottom_top, soil_layers
Data variables:
ACGRDFLX (time, south_north, west_east) float32 0.0 ...
ACHFX (time, south_north, west_east) float32 173706.0 ...
ACLHF (time, south_north, west_east) float32 5.62606e+06 ...
ALBBCK (time, south_north, west_east) float32 0.08 ...
ALBEDO (time, south_north, west_east) float32 0.08 ...
AVG_FUEL_FRAC (time, south_north, west_east) float32 0.0 ...
CANWAT (time, south_north, west_east) float32 0.0 ...
CF1 (time) float32 1.76177 1.76177 ...
CF2 (time) float32 -1.02047 -1.02047 ...
CF3 (time) float32 0.258698 0.258698 ...
CFN (time) float32 1.46268 1.46268 ...
CFN1 (time) float32 -0.462685 -0.462685 ...
CLDFRA (time, bottom_top, south_north, west_east) float32 0.0 ...
COSALPHA (south_north, west_east) float32 0.975546 ...
DN (bottom_top) float32 0.0 ...
DNW (bottom_top) float32 -0.00700003 ...
DZS (soil_layers) float32 0.2 0.45 0.8
E (south_north, west_east) float32 0.000145782 ...
EL_PBL (time, bottom_top, south_north, west_east) float32 0.0 ...
EMISS (time, south_north, west_east) float32 0.98 ...
F (south_north, west_east) float32 4.12304e-06 ...
FNM (bottom_top) float32 0.0 0.411766 ...
FNP (bottom_top) float32 0.0 0.588234 ...
GLW (time, south_north, west_east) float32 404.028 ...
GRAUPELNC (time, south_north, west_east) float32 0.0 ...
GRDFLX (time, south_north, west_east) float32 0.0 ...
HAILNC (time, south_north, west_east) float32 0.0 ...
HFX (time, south_north, west_east) float32 4.29665 ...
HFX_FORCE (time) float32 0.0 0.0 0.0 0.0 0.0 ...
HFX_FORCE_TEND (time) float32 0.0 0.0 0.0 0.0 0.0 ...
HGT (south_north, west_east) float32 0.0 ...
H_DIABATIC (time, bottom_top, south_north, west_east) float32 0.0 ...
ISLTYP (south_north, west_east) int32 14 ...
ITIMESTEP (time) int32 288 360 432 504 576 ...
IVGTYP (south_north, west_east) int32 16 ...
K22_SHALLOW (time, south_north, west_east) int32 0 ...
KBCON_SHALLOW (time, south_north, west_east) int32 0 ...
KTOP_SHALLOW (time, south_north, west_east) int32 0 ...
LAI (south_north, west_east) float32 0.0 ...
LANDMASK (south_north, west_east) float32 0.0 ...
LH (time, south_north, west_east) float32 140.399 ...
LH_FORCE (time) float32 0.0 0.0 0.0 0.0 0.0 ...
LH_FORCE_TEND (time) float32 0.0 0.0 0.0 0.0 0.0 ...
LU_INDEX (south_north, west_east) float32 16.0 ...
MAPFAC_M (south_north, west_east) float32 1.14636 ...
MAPFAC_MX (south_north, west_east) float32 1.14636 ...
MAPFAC_MY (south_north, west_east) float32 1.14636 ...
MAPFAC_U (south_north, west_east) float32 1.14637 ...
MAPFAC_UX (south_north, west_east) float32 1.14637 ...
MAPFAC_UY (south_north, west_east) float32 1.14637 ...
MAPFAC_V (south_north, west_east) float32 1.14637 ...
MAPFAC_VX (south_north, west_east) float32 1.14637 ...
MAPFAC_VY (south_north, west_east) float32 1.14637 ...
MAX_MSTFX (time) float32 0.0 0.0 0.0 0.0 0.0 ...
MAX_MSTFY (time) float32 0.0 0.0 0.0 0.0 0.0 ...
MF_VX_INV (south_north, west_east) float32 0.872322 ...
MU (time, south_north, west_east) float32 540.09 ...
MUB (time, south_north, west_east) float32 95000.0 ...
NEST_POS (south_north, west_east) float32 0.0 ...
NOAHRES (time, south_north, west_east) float32 0.0 ...
OLR (time, south_north, west_east) float32 285.326 ...
P (time, bottom_top, south_north, west_east) float32 951.938 ...
P00 (time) float32 100000.0 100000.0 ...
PB (time, bottom_top, south_north, west_east) float32 99667.5 ...
PBLH (time, south_north, west_east) float32 803.482 ...
PH (time, bottom_top, south_north, west_east) float32 17.5665 ...
PHB (time, bottom_top, south_north, west_east) float32 277.505 ...
POTEVP (time, south_north, west_east) float32 0.0 ...
PSFC (time, south_north, west_east) float32 100959.0 ...
P_HYD (time, bottom_top, south_north, west_east) float32 100618.0 ...
P_TOP (time) float32 5000.0 5000.0 5000.0 ...
Q2 (time, south_north, west_east) float32 0.0201797 ...
QCLOUD (time, bottom_top, south_north, west_east) float32 0.0 ...
QFX (time, south_north, west_east) float32 5.61596e-05 ...
QGRAUP (time, bottom_top, south_north, west_east) float32 0.0 ...
QICE (time, bottom_top, south_north, west_east) float32 0.0 ...
QNICE (time, bottom_top, south_north, west_east) float32 0.0 ...
QNRAIN (time, bottom_top, south_north, west_east) float32 0.0 ...
QRAIN (time, bottom_top, south_north, west_east) float32 0.0 ...
QSNOW (time, bottom_top, south_north, west_east) float32 0.0 ...
QVAPOR (time, bottom_top, south_north, west_east) float32 0.0168769 ...
RAINC (time, south_north, west_east) float32 0.0 ...
RAINNC (time, south_north, west_east) float32 0.0 ...
RAINSH (time, south_north, west_east) float32 0.0 ...
RDN (bottom_top) float32 0.0 -117.647 ...
RDNW (bottom_top) float32 -142.857 ...
RDX (time) float32 3.33333e-05 ...
RDY (time) float32 3.33333e-05 ...
RESM (time) float32 0.0 0.0 0.0 0.0 0.0 ...
SAVE_TOPO_FROM_REAL (time) int32 0 0 0 0 0 0 0 0 0
SEAICE (time, south_north, west_east) float32 0.0 ...
SEED1 (time) int32 0 0 0 0 0 0 0 0 0
SEED2 (time) int32 0 0 0 0 0 0 0 0 0
SFROFF (time, south_north, west_east) float32 0.0 ...
SH2O (time, soil_layers, south_north, west_east) float32 1.0 ...
SINALPHA (south_north, west_east) float32 0.219797 ...
SMCREL (time, soil_layers, south_north, west_east) float32 1.0 ...
SMOIS (time, soil_layers, south_north, west_east) float32 1.0 ...
SNOPCX (time, south_north, west_east) float32 0.0 ...
SNOW (time, south_north, west_east) float32 0.0 ...
SNOWC (time, south_north, west_east) float32 0.0 ...
SNOWH (time, south_north, west_east) float32 0.0 ...
SNOWNC (time, south_north, west_east) float32 0.0 ...
SOILTB (time, south_north, west_east) float32 0.0 ...
SR (time, south_north, west_east) float32 0.0 ...
SST (time, south_north, west_east) float32 302.047 ...
SSTSK (time, south_north, west_east) float32 0.0 ...
STEPAVE_COUNT (time) int32 0 0 0 0 0 0 0 0 0
SWDOWN (time, south_north, west_east) float32 0.0 ...
SWNORM (time, south_north, west_east) float32 0.0 ...
T (time, bottom_top, south_north, west_east) float32 0.658561 ...
T00 (time) float32 290.0 290.0 290.0 ...
T2 (time, south_north, west_east) float32 301.709 ...
TH2 (time, south_north, west_east) float32 300.887 ...
TISO (time) float32 0.0 0.0 0.0 0.0 0.0 ...
TKE_PBL (time, bottom_top, south_north, west_east) float32 0.105927 ...
TLP (time) float32 50.0 50.0 50.0 50.0 ...
TMN (time, south_north, west_east) float32 302.265 ...
TSK (time, south_north, west_east) float32 302.047 ...
TSK_FORCE (time) float32 0.0 0.0 0.0 0.0 0.0 ...
TSK_FORCE_TEND (time) float32 0.0 0.0 0.0 0.0 0.0 ...
TSLB (time, soil_layers, south_north, west_east) float32 287.603 ...
U (time, bottom_top, south_north, west_east) float32 -1.96133 ...
U10 (time, south_north, west_east) float32 -1.86241 ...
UAH (time, south_north, west_east) float32 0.0 ...
UDROFF (time, south_north, west_east) float32 0.0 ...
UST (time, south_north, west_east) float32 0.20173 ...
V (time, bottom_top, south_north, west_east) float32 6.60783 ...
V10 (time, south_north, west_east) float32 6.27481 ...
VAH (time, south_north, west_east) float32 0.0 ...
VEGFRA (south_north, west_east) float32 0.0 ...
W (time, bottom_top, south_north, west_east) float32 -0.000252068 ...
XICEM (time, south_north, west_east) float32 0.0 ...
XLAND (time, south_north, west_east) float32 2.0 ...
XMB_SHALLOW (time, south_north, west_east) float32 0.0 ...
xtime (time) float32 720.0 900.0 1080.0 ...
ZETATOP (time) float32 0.0 0.0 0.0 0.0 0.0 ...
ZNU (time, bottom_top) float32 0.9965 ...
ZNW (time, bottom_top) float32 0.9965 ...
ZS (soil_layers) float32 0.15 0.475 1.1
WS (time, bottom_top, south_north, west_east) float32 6.89276 ...
GEOPOTENTIAL (time, bottom_top, south_north, west_east) float32 295.071 ...
SLP (time, south_north, west_east) float32 1009.71 ...
Z (time, bottom_top, south_north, west_east) float32 30.0786 ...
T2C (time, south_north, west_east) float32 28.5587 ...
PRESSURE (time, bottom_top, south_north, west_east) float32 100619.0 ...
THETA (time, bottom_top, south_north, west_east) float32 300.659 ...
TK (time, bottom_top, south_north, west_east) float32 301.19 ...
PRCP (time, south_north, west_east) float32 nan ...
PRCP_NC (time, south_north, west_east) float32 nan ...
PRCP_C (time, south_north, west_east) float32 nan ...
Attributes:
TITLE: OUTPUT FROM WRF V3.3.1 MODEL
START_DATE: 2005-09-21_00:00:00
SIMULATION_START_DATE: 2005-09-20_12:00:00
WEST-EAST_GRID_DIMENSION: 201
SOUTH-NORTH_GRID_DIMENSION: 201
BOTTOM-TOP_GRID_DIMENSION: 28
DX: 30000.0
DY: 30000.0
GRIDTYPE: C
DIFF_OPT: 1
KM_OPT: 4
DAMP_OPT: 3
DAMPCOEF: 0.2
KHDIF: 0.0
KVDIF: 0.0
MP_PHYSICS: 8
RA_LW_PHYSICS: 1
RA_SW_PHYSICS: 1
SF_SFCLAY_PHYSICS: 2
SF_SURFACE_PHYSICS: 2
BL_PBL_PHYSICS: 2
CU_PHYSICS: 5
SURFACE_INPUT_SOURCE: 1
SST_UPDATE: 1
GRID_FDDA: 0
GFDDA_INTERVAL_M: 0
GFDDA_END_H: 0
GRID_SFDDA: 0
SGFDDA_INTERVAL_M: 0
SGFDDA_END_H: 0
SF_URBAN_PHYSICS: 0
FEEDBACK: 1
SMOOTH_OPTION: 0
SWRAD_SCAT: 1.0
W_DAMPING: 0
MOIST_ADV_OPT: 1
SCALAR_ADV_OPT: 1
TKE_ADV_OPT: 1
DIFF_6TH_OPT: 2
DIFF_6TH_FACTOR: 0.12
OBS_NUDGE_OPT: 0
BUCKET_MM: -1.0
BUCKET_J: -1.0
PREC_ACC_DT: 0.0
OMLCALL: 0
ISFTCFLX: 0
ISHALLOW: 0
DFI_OPT: 0
SHCU_PHYSICS: 0
WEST-EAST_PATCH_START_UNSTAG: 1
WEST-EAST_PATCH_END_UNSTAG: 200
WEST-EAST_PATCH_START_STAG: 1
WEST-EAST_PATCH_END_STAG: 201
SOUTH-NORTH_PATCH_START_UNSTAG: 1
SOUTH-NORTH_PATCH_END_UNSTAG: 200
SOUTH-NORTH_PATCH_START_STAG: 1
SOUTH-NORTH_PATCH_END_STAG: 201
BOTTOM-TOP_PATCH_START_UNSTAG: 1
BOTTOM-TOP_PATCH_END_UNSTAG: 27
BOTTOM-TOP_PATCH_START_STAG: 1
BOTTOM-TOP_PATCH_END_STAG: 28
GRID_ID: 1
PARENT_ID: 1
I_PARENT_START: 1
J_PARENT_START: 1
PARENT_GRID_RATIO: 1
DT: 150.0
CEN_LAT: 30.0
CEN_LON: 87.0
TRUELAT1: 30.0
TRUELAT2: 35.0
MOAD_CEN_LAT: 30.0
STAND_LON: 87.0
POLE_LAT: 90.0
POLE_LON: 0.0
GMT: 12.0
JULYR: 2005
JULDAY: 263
MAP_PROJ: 1
MMINLU: USGS
NUM_LAND_CAT: 28
ISWATER: 16
ISLAKE: 28
ISICE: 24
ISURBAN: 1
ISOILWATER: 14
history: Thu Nov 10 22:29:11 CET 2011: runncl.sh /cfs/tool/jc_1.4test/utils//POST_wrf.ncl ./wrfout_d01_2005-09-21_00-00-00_24h.nc . 0 vars_stat.csv vars_del.csv
Thu Nov 10 22:29:08 2011: ncks -d Time,4,12 wrfout_d01_2005-09-20_12:00:00 wrfout_d01_2005-09-21_00-00-00_24h.nc
NCO: 20111110
CREATION_DATE: Thu Nov 10 22:29:11 CET 2011
SPINUP_INDEX: 0
END_INDEX: 8
pyproj_srs: +units=m +proj=lcc +lat_1=30.0 +lat_2=35.0 +lat_0=30.0 +lon_0=87.0 +x_0=0 +y_0=0 +a=6370000 +b=6370000
Diagnostic variables¶
wrf.T2C.mean(dim='time', keep_attrs=True).salem.quick_map();
3D interpolation¶
ws_h = wrf.isel(time=5).salem.wrf_zlevel('WS', levels=10000.)
ws_h.salem.quick_map(cmap='Reds');
Final remarks¶
- xarray relies on pandas, which is one of the most widely used scientific python tools
- their documentation is excellent
- both libraries require a certain learning investment, but this time is well spent
- there is potential for "ACINN homegrown" tools based on these libs
