Numpy, part I: why numpy?¶
You might have read somewhere that Python is "slow" in comparison to other languages like Fortran or C. While generally true, this statement has only little meaning without context. As a scripting language, python is fast enough. For numerical computations (like the computations done by an atmospheric model or by a machine learning algorithm), pure Python is quite slow indeed. Fortunately, there are ways to overcome this problem!
In this chapter we are going to explain why the numpy library was created. Before that, we will discuss some of the differences between python and compiled languages widely used in scientific software development (like C and FORTRAN).
Table of Contents
Why is python "slow"?¶
Last week, we've learned that the memory consumption of a python int is larger than the memory needed to store the binary number alone. This overhead in memory consumption is due to the nature of python data types, which are all objects. We've already learned that these objects come with certain "services", for example:
s = 'this.needs.some.splitting'
' '.join(s.split('.')).title() # join, split, title are all "functions" applyed to a string
There is not a single thing in the python language which isn't an object. Yes, even functions are objects! Let me prove it to you:
def useful_function(a, b):
"""This function adds two objects together.
Parameters
----------
a : an object
b : another object
Returns
-------
The sum of the two
"""
return a + b
type(useful_function)
print(useful_function.__doc__)
Functions are objects of type function, and one of their attributes (__doc__) gives us access to their docstring. During the course of the semester you are going to learn how to use more and more of these object features, and hopefully you are going to like them more and more (at least this is what happened to me).
Now, why does this make python "slow"? Well, in simple words, these "services" tend to increase the complexity and the number of operations an interpreter has to perform when running a program. More specialized languages will be less flexible than python, but will be faster at running specialized operations and be less memory hungry.
Python's high-level of abstraction (i.e. python's flexibility) makes it slower than its lower-level counterparts like C or FORTRAN. But, why is that so?
Dynamically versus statically typed languages¶
Python is a so-called dynamically typed language, which means that the type of a variable is determined by the interpreter at run time. To understand what that means in practice, let's have a look at the following code snippet:
a = 2
b = 3
c = a + b
The line c = a + b is valid python syntax. The operation that has to be applied by the + operator, however, depends on the type of the variables to be added. Remember what happens when adding two lists for example:
a = [2]
b = [3]
a + b
In this simple example it would be theoretically possible for the interpreter to predict which operation to apply beforehand (by prior parsing of all lines of the cell). In most cases, however, this is impossible (think of the jupyter notebook cells for example, which can be executed in any order).
Languages which assess the type of variables at run time are called dynamically typed programming languages. Matlab, Python or R are examples falling in this category.
Statically typed languages, however, require the programmer to provide the type of variables while writing the code. Here is an example of a program written in C:
#include <stdio.h>
int main ()
{
int a = 2;
int b = 3;
int c = a + b;
printf ("Sum of two numbers : %d \n", c);
}
The variable type definition is part of the C syntax. This applies to the variables themselves, but also to the output of computations. This is a fundamental difference to python, and comes with several advantages. Static typing usually results in code that executes faster: since the program knows the exact data types that are in use, it can predict the memory consumption of operations beforehand and produce optimized machine code. Another advantage is code documentation: the statement int c = a + b makes it clear that we are adding two numbers while the python equivalent c = a + b could produce a number, a string, a list, etc.
Compiled versus interpreted languages¶
Statically typed languages often require compilation. To run the C code snippet I had to create a new text file (example.c), write the code, compile it ($ gcc -o myprogram example.c), before finally being able to execute it ($ ./myprogram).
gcc is the compiler I used to translate the C source code (a text file) to a low level language (machine code) in order to create an executable (myprogram). Later changes to the source code require a new compilation step for the changes to take effect.
Because of this "edit-compile-run" cycle, compiled languages are not interactive: there is no equivalent to python's command line interpreter in C. Compiling complex programs can take from a couple of minutes to several hours in some extreme cases. This compilation time, however, is usually associated with faster execution times: as mentioned earlier, the compiler's task is to optimize the program for memory consumption by source code analysis. Often, a compiled program is optimized for the machine architecture it is compiled onto. Like interpreters, there can be different compilers for the same language. They differ in the optimization steps they undertake to make the program faster, and in their support of various hardware architectures.
Interpreters do not require compilation: they analyze the code at run time. The following code for example is syntactically correct:
def my_func():
a = 1
b = '2'
return a + b
but its execution of this code results in a TypeError:
my_func()
The interpreter cannot detect these errors before runtime.
Parenthesis I: python bytecode
When executing a python program from the command line, the CPython interpreter creates a hidden directory called __pycache__. This directory contains bytecode files, which are your python source code files translated to binary files. This is an optimization step which makes subsequent executions of the program run faster. While this conversion step is sometimes called "compilation", it should not be mistaken with a C-program compilation: indeed, python bytecode still needs an interpreter to run, while compiled executables can be run without C interpreter.
Parenthesis II: static typing and compilation
Statically typed languages are often compiled, and dynamically typed languages are often interpreted. While this is a good general rule of thumb, this is not always true and the vast landscape of programming languages contains many exceptions. This lecture is only a very short introduction to these concepts: you'll have to refer to more advanced computer science lectures if you want to learn about these topics in more detail.
Here comes numpy¶
Let's summarize the two previous chapters:
- Python is flexible, interactive and slow
- C is less flexible, non-interactive and fast
This is an oversimplification, but a quite realistic one. On top of that, python's built-in list data types are far from what scientists need to do arithmetics or vector computations. To add two lists (element-wise), one must write:
def add_lists(A, B):
"""Element-wise addition of two lists."""
return [a+b for a, b in zip(A, B)]
add_lists([1, 2, 3], [4, 5, 6])
The numpy equivalent is much more intuitive and straightforward:
import numpy as np
def add_arrays(A, B):
return np.add(A, B)
add_arrays([1, 2, 3], [4, 5, 6])
 Question: why did I prefer to use the
Question: why did I prefer to use the np.add function here instead of the simpler + operator?
Let's see which of the two functions runs faster:
n = 10
A = np.random.randn(n)
B = np.random.randn(n)
%timeit add_lists(A, B)
%timeit add_arrays(A, B)
Numpy is approximately 5 times faster.
 Exercise: repeat the performance test with n=100 and n=10000. How does the performance scale with the size of the array? Now repeat the test but make the input arguments
Exercise: repeat the performance test with n=100 and n=10000. How does the performance scale with the size of the array? Now repeat the test but make the input arguments A and B lists instead of numpy arrays. How is the performance comparison in this case? Why?
Why is numpy so much faster than pure python? One of the major reasons is vectorization, which is the process of applying mathematical operations to all elements of an array ("vector") at once instead of looping through them like we would do in pure python. "for loops" in python are slow because for each addition, python has to:
- access the elements a and b in the list A and B
- check the type of both a and b
- apply the
+operator on the data they store - store the result.
Numpy skips the first two steps and does them only once before the actual operation. What does numpy know about the addition operation that the pure python version can't infer?
- the type of all numbers to add
- the type of the output
- the size of the output array (same as input)
Numpy can use this information to optimize the computation, but this isn't possible without trade-offs. See the following for example:
add_lists([1, 'foo'], [3, 'bar']) # works fine
add_arrays([1, 'foo'], [3, 'bar']) # raises a TypeError
$\rightarrow$ numpy can only be that fast because the input and output data types are uniform and known before the operation.
Internally, numpy achieves vectorization by relying on a lower-level, statically typed and compiled language: C! At the time of writing, 55% of the numpy codebase is written in C/C++. The rest of the codebase offers an interface (a "layer") between python and the internal C code.
As a result, numpy has to be compiled at installation. Most users do not notice this compilation step anymore (recent pip and conda installations are shipped with pre-compiled binaries), but installing numpy used to require several minutes on my laptop when I started to learn python myself.
Anatomy of a ndarray¶
From the numpy reference:
N-dimensional ndarray's are the core structure of the numpy library. It is a multidimensional container of items of the same type and size. The number of dimensions and items in an array is defined by its shape, which is a tuple of N positive integers that specify the number of items in each dimension. The type of items in the array is specified by a separate data-type object (dtype), one of which is associated with each ndarray.
All ndarrays are homogenous: every item takes up the same size block of memory, and all blocks are interpreted in exactly the same way.
An item extracted from an array, e.g., by indexing, is represented by a Python object whose type is one of the array scalar types built in NumPy.
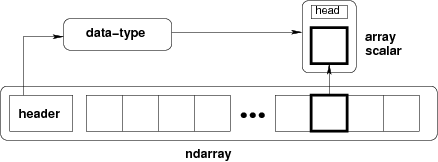
Figure: Conceptual diagram showing the relationship between the three fundamental objects used to describe the data in an array: 1) the ndarray itself, 2) the data-type object that describes the layout of a single fixed-size element of the array, 3) the array-scalar Python object that is returned when a single element of the array is accessed.
This contiguous one-dimensional segment of computer memory stored by the array is combined with an indexing scheme that maps N-dimensional indexes into the location of an item in the block. Concretely, this means that there is no difference in the memory layout of these two arrays:
a = np.arange(9)
b = a.reshape((3, 3))
Both are internally stored using a one dimensional memory block. The difference lies in the way numpy gives access to the internal data:
a[4], b[1, 1]
This means that elementwise operations have the same execution speed for N dimensional arrays as for 1-D arrays. Now, let's replace an element from array b:
b[1, 1] = 99
b
Since the indexing [1, 1] is on the left-hand side of the assignment operator, we modified b in-place. Numpy located the block of memory that needed to be changed and replaced it with another number. Since all these numbers are integers with a fixed memory size, this is not a problem. But what happens if we try to assign another data type?
b[1, 1] = 999.99
b
The float is converted to an integer! This is a dangerous "feature" of numpy and should be used with care. Another extremely important mechanism of ndarrays is their internal handling of data. Indeed:
a
What happened here? Modifying the data in b also modified the data in a. More precisely, both arrays share the same internal data: b is a view of the data owned by a.
b.base is a
a.base is None
np.shares_memory(a, b)
This allows for memory efficient reshaping and vector operations on numpy arrays, but is a source of confusion for many. Next week we will look into more detail which operations return a view and which return a copy.
Take home points¶
- The process of type checking may occur either at compile-time (statically typed language) or at runtime (dynamically typed language). These terms are not usually used in a strict sense.
- Statically typed languages are often compiled, while dynamically typed languages are interpreted.
- There is a trade-off between the flexibility of a language and its speed: static typing allows programs to be optimized at compilation time, thus allowing them to run faster.
- When speed matters, python allows to use compiled libraries under a python interface. numpy is using C under the hood to optimize its computations.
- numpy arrays use a continuous block of memory of homogenous data type. This allows for faster memory access and easy vectorization of mathematical operations.
Further reading¶
I highly recommend to have a look at the first part of Jake Vanderplas' blog post, Why python is slow (up to the "hacking" part). It provides more details and a good visual illustration of the c = a + b example. The second part is a more involved read, but very interesting too!
Addendum: is python really that slow?¶
The short answer is: yes, python is slower than a number of other languages. You'll find many benchmarks online illustrating it.
Is it bad?
No. Jake Vanderplas (a well known contributor of the scientific python community) writes:
As well, it comes down to this: dynamic typing makes Python easier to use than C. It's extremely flexible and forgiving, this flexibility leads to efficient use of development time, and on those occasions that you really need the optimization of C or Fortran, Python offers easy hooks into compiled libraries. It's why Python use within many scientific communities has been continually growing. With all that put together, Python ends up being an extremely efficient language for the overall task of doing science with code.
It's the flexibility and readability of python that makes it so popular. Python is the language of choice for major actors like instagram or spotify, and is has become the high-level interface to highly optimized machine learning libraries like TensorFlow or Torch.
For a scientist, writing code efficiently is much more important than writing efficient code. Or is it?
What's next?¶
Back to the table of contents, or jump to this week's assignment.